MAPK6
| MAPK6 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | MAPK6, ERK3, HsT17250, PRKM6, p97MAPK, mitogen-activated protein kinase 6 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 602904; MGI: 1354946; HomoloGene: 55683; GeneCards: MAPK6; OMA:MAPK6 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Mitogen-activated protein kinase 6 is an enzyme that in humans is encoded by the MAPK6 gene.[5][6]
The protein encoded by this gene is a member of the Ser/Thr protein kinase family, and is most closely related to mitogen-activated protein kinases (MAP kinases). MAP kinases, also known as extracellular signal-regulated kinases (ERKs), are activated through protein phosphorylation cascades and act as integration points for multiple biochemical signals. This kinase is localized in the nucleus, and has been reported to be activated in fibroblasts upon treatment with serum or phorbol esters.[6]
Discovery
ERK3/MAPK6 was initially cloned from the rat brain cDNA library by homology screening with probes ERK1 derived probe.[7]
Gene location
In humans, MAPK 6 gene is located on the distal arm of chromosome 15 (15q21.2). It is 47.01kb long and is transcribed in the centromere to telomere orientation. It consist of 6 exons with the translation initiation codon which is located in exon2.[8]
Structure
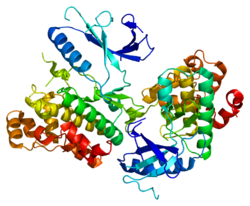
It is an atypical member of the mitogen activated kinases family. The molecular mass of the translated protein is approximately 100kDa, and is made up of 721 amino acid residues.[8][7] It contains a typical kinase domain at the N- terminal and an extended C- terminal. The first 150 residues at c- terminal are 50% similar to ERK4 protein. At the kinase domain it exhibits about 70% similarity with the ERK4 protein.[8][7] The activation loop of the phosphorylation motif contains only one phospho acceptor site (Ser-Glu-Gly).[7]
The structure is predicted by homology modelling using the crystal structure of phoshphorylated ERK2. According to the model, the structure of ERK3/MAPK6 kinase domain resembles other MAP kinases. The modelled ERK3/MAPK6 kinase domain is predicted to fold with a topology similar to other MAP kinases.[7]
Expression
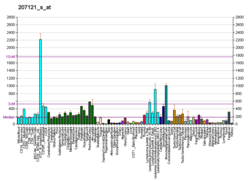
ERK3/MAPK6 is widely expressed protein however it is expressed in significantly higher amounts in skeletal muscles and brain. It is localized in cytoplasm and the nucleus of cells. ERK3/MAPK6 is a highly unstable protein and has a very little half life of less than an hour. It is degraded by ubiquitin mediated proteasomal pathway.[8]
Function
It is very important for neonatal growth and survival. ERK3/MAPK6 forms a complex with microtubule associated protein2 (MAP2) and MAPKAPK5 which mediates the phosphorylation of MAPKAPK5 which in turn phosphorylates ERK3/MAPK6 at serine 189 residue mediating the entry into cell cycle.[9] It also acts as a regulator for T- cell development. The catalytic activity of ERK3/MAPK6 plays an important for the proper differentiation of T-cells in the thymus. The long c- terminal is responsible for thymic differentiation.[10]
Role in cancer
ERK3/MAPK6 interacts with and phosphorylated steroid receptor coactivator 3 (SRC-3) This coreceptor is an oncogenic protein which when overexpressed at serine 857 leads to cancer. After the phosphorylation of SRC-3 results in the upregulation of MMP activity ERK3-mediated phosphorylation at S857 was essential for interaction of SRC-3 with the ETS transcription factor PEA3, which promotes upregulation of MMP gene expression and proinvasive activity.[11]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000069956 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000042688 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Meloche S (2005-04-01). "Erk3". AfCS-Nature Molecule Pages. doi:10.1038/mp.a000876.01 (inactive 2024-04-11). ISSN 1477-5921.
{{cite journal}}: CS1 maint: DOI inactive as of April 2024 (link) - ^ a b "MAPK6 mitogen-activated protein kinase 6 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2018-11-09.
- ^ a b c d e Coulombe P, Meloche S (August 2007). "Atypical mitogen-activated protein kinases: structure, regulation and functions". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1773 (8): 1376–87. doi:10.1016/j.bbamcr.2006.11.001. PMID 17161475.
- ^ a b c d "MAPK6 (mitogen-activated protein kinase 6)". atlasgeneticsoncology.org. Retrieved 2018-11-09.
- ^ "Volume II Accession Number Index", Rodents, Elsevier, 1987, pp. ACCESSION–1–ACCESSION–5, doi:10.1016/b978-0-12-512512-3.50015-8, ISBN 978-0-12-512512-3
- ^ Marquis M, Daudelin JF, Boulet S, Sirois J, Crain K, Mathien S, Turgeon B, Rousseau J, Meloche S, Labrecque N (September 2014). "The catalytic activity of the mitogen-activated protein kinase extracellular signal-regulated kinase 3 is required to sustain CD4+ CD8+ thymocyte survival". Molecular and Cellular Biology. 34 (18): 3374–87. doi:10.1128/MCB.01701-13. PMC 4135614. PMID 25002529.
- ^ Long W, Foulds CE, Qin J, Liu J, Ding C, Lonard DM, Solis LM, Wistuba II, Qin J, Tsai SY, Tsai MJ, O'Malley BW (May 2012). "ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion". The Journal of Clinical Investigation. 122 (5): 1869–80. doi:10.1172/jci61492. PMC 3336992. PMID 22505454.
Further reading
- Boulton TG, Nye SH, Robbins DJ, Ip NY, Radziejewska E, Morgenbesser SD, DePinho RA, Panayotatos N, Cobb MH, Yancopoulos GD (May 1991). "ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF". Cell. 65 (4): 663–75. doi:10.1016/0092-8674(91)90098-J. PMID 2032290. S2CID 35051321.
- Zhu AX, Zhao Y, Moller DE, Flier JS (December 1994). "Cloning and characterization of p97MAPK, a novel human homolog of rat ERK-3". Molecular and Cellular Biology. 14 (12): 8202–11. doi:10.1128/MCB.14.12.8202. PMC 359359. PMID 7969157.
- Cheng M, Boulton TG, Cobb MH (April 1996). "ERK3 is a constitutively nuclear protein kinase". The Journal of Biological Chemistry. 271 (15): 8951–8. doi:10.1074/jbc.271.15.8951. PMID 8621539.
- Sauma S, Friedman E (May 1996). "Increased expression of protein kinase C beta activates ERK3". The Journal of Biological Chemistry. 271 (19): 11422–6. doi:10.1074/jbc.271.19.11422. PMID 8626698.
- Zimmermann J, Lamerant N, Grossenbacher R, Furst P (April 2001). "Proteasome- and p38-dependent regulation of ERK3 expression". The Journal of Biological Chemistry. 276 (14): 10759–66. doi:10.1074/jbc.M008567200. PMID 11148204.
- Robinson MJ, Xu Be BE, Stippec S, Cobb MH (February 2002). "Different domains of the mitogen-activated protein kinases ERK3 and ERK2 direct subcellular localization and upstream specificity in vivo". The Journal of Biological Chemistry. 277 (7): 5094–100. doi:10.1074/jbc.M110935200. PMID 11741894.
- Kinet S, Bernard F, Mongellaz C, Perreau M, Goldman FD, Taylor N (October 2002). "gp120-mediated induction of the MAPK cascade is dependent on the activation state of CD4(+) lymphocytes". Blood. 100 (7): 2546–53. doi:10.1182/blood-2002-03-0819. PMID 12239168.
- Coulombe P, Rodier G, Pelletier S, Pellerin J, Meloche S (July 2003). "Rapid turnover of extracellular signal-regulated kinase 3 by the ubiquitin-proteasome pathway defines a novel paradigm of mitogen-activated protein kinase regulation during cellular differentiation". Molecular and Cellular Biology. 23 (13): 4542–58. doi:10.1128/MCB.23.13.4542-4558.2003. PMC 164847. PMID 12808096.
- Julien C, Coulombe P, Meloche S (October 2003). "Nuclear export of ERK3 by a CRM1-dependent mechanism regulates its inhibitory action on cell cycle progression". The Journal of Biological Chemistry. 278 (43): 42615–24. doi:10.1074/jbc.M302724200. PMID 12915405.
- Rai R, Mahale A, Saranath D (August 2004). "Molecular cloning, isolation and characterisation of ERK3 gene from chewing-tobacco induced oral squamous cell carcinoma". Oral Oncology. 40 (7): 705–12. doi:10.1016/j.oraloncology.2004.01.010. PMID 15172640.
- Coulombe P, Rodier G, Bonneil E, Thibault P, Meloche S (July 2004). "N-Terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome". Molecular and Cellular Biology. 24 (14): 6140–50. doi:10.1128/MCB.24.14.6140-6150.2004. PMC 434260. PMID 15226418.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- Hoeflich KP, Eby MT, Forrest WF, Gray DC, Tien JY, Stern HM, Murray LJ, Davis DP, Modrusan Z, Seshagiri S (October 2006). "Regulation of ERK3/MAPK6 expression by BRAF". International Journal of Oncology. 29 (4): 839–49. doi:10.3892/ijo.29.4.839. PMID 16964379.
External links
- MAP Kinase Resource Archived 2021-04-15 at the Wayback Machine.
- v
- t
- e
-
 2i6l: Crystal structure of human mitogen activated protein kinase 6 (MAPK6)
2i6l: Crystal structure of human mitogen activated protein kinase 6 (MAPK6)