ATP5F1A
| ATP5F1A | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ATP5F1A, ATP5A, ATP5AL2, ATPM, HEL-S-123m, MC5DN4, MOM2, OMR, ORM, hATP1, COXPD22, ATP synthase, H+ transporting, mitochondrial F1 complex, alpha 1, ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle, ATP5A1, ATP synthase F1 subunit alpha | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 164360 MGI: 88115 HomoloGene: 2985 GeneCards: ATP5F1A | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
ATP synthase F1 subunit alpha, mitochondrial is an enzyme that in humans is encoded by the ATP5F1A gene.[5][6]
Function
This gene encodes a subunit of mitochondrial ATP synthase. Mitochondrial ATP synthase catalyzes ATP synthesis, using an electrochemical gradient of protons across the inner membrane during oxidative phosphorylation. ATP synthase is composed of two linked multi-subunit complexes: the soluble catalytic core, F1, and the membrane-spanning component, Fo, comprising the proton channel. The catalytic portion of mitochondrial ATP synthase consists of 5 different subunits (alpha, beta, gamma, delta, and epsilon) assembled with a stoichiometry of 3 alpha, 3 beta, and a single representative of the other 3. The proton channel consists of three main subunits (a, b, c). This gene encodes the alpha subunit of the catalytic core. Alternatively spliced transcript variants encoding the same protein have been identified. Pseudogenes of this gene are located on chromosomes 9, 2, and 16.[6]
Structure
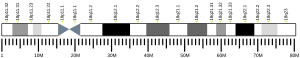
The ATP5F1A gene, located on the q arm of chromosome 18 in position 21, is made up of 13 exons and is 20,090 base pairs in length.[6] The ATP5F1A protein weighs 59.7 kDa and is composed of 553 amino acids.[7][8] The protein is a subunit of the catalytic portion of the F1Fo ATPase, also known as Complex V, which consists of 14 nuclear and 2 mitochondrial -encoded subunits. As an alpha subunit, ATP5F1A is contained within the catalytic F1 portion of the complex.[6] The nomenclature of the enzyme has a long history. The F1 fraction derives its name from the term "Fraction 1" and Fo (written as a subscript letter "o", not "zero") derives its name from being the binding fraction for oligomycin, a type of naturally-derived antibiotic that is able to inhibit the Fo unit of ATP synthase.[9][10] The F1 particle is large and can be seen in the transmission electron microscope by negative staining.[11] These are particles of 9 nm diameter that pepper the inner mitochondrial membrane. They were originally called elementary particles and were thought to contain the entire respiratory apparatus of the mitochondrion, but, through a long series of experiments, Efraim Racker and his colleagues (who first isolated the F1 particle in 1961) were able to show that this particle is correlated with ATPase activity in uncoupled mitochondria and with the ATPase activity in submitochondrial particles created by exposing mitochondria to ultrasound. This ATPase activity was further associated with the creation of ATP by a long series of experiments in many laboratories.
Function
Mitochondrial membrane ATP synthase (F1Fo ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F1 - containing the extramembraneous catalytic core, and Fo - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F1 is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Subunits alpha and beta form the catalytic core in F1. Rotation of the central stalk against the surrounding alpha(3)beta(3) subunits leads to hydrolysis of ATP in three separate catalytic sites on the beta subunits. Subunit alpha does not bear the catalytic high-affinity ATP-binding sites.[12]
Clinical significance
Mutations affecting the ATP5F1A gene cause combined oxidative phosphorylation deficiency 22 (COXPD22), a mitochondrial disorder characterized by intrauterine growth retardation, microcephaly, hypotonia, pulmonary hypertension, failure to thrive, encephalopathy, and heart failure. Mutations on the ATP5F1A gene also cause mitochondrial complex V deficiency, nuclear 4 (MC5DN4), a mitochondrial disorder with heterogeneous clinical manifestations including dysmorphic features, psychomotor retardation, hypotonia, growth retardation, cardiomyopathy, enlarged liver, hypoplastic kidneys and elevated lactate levels in urine, plasma and cerebrospinal fluid.[13]
Resveratrol inhibition of the F1 catalytic core increases adenosine monophosphate (AMP) levels, thereby activating the AMP-activated protein kinase enzyme.[14]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000152234 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000025428 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Kataoka H, Biswas C (July 1991). "Nucleotide sequence of a cDNA for the alpha subunit of human mitochondrial ATP synthase". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1089 (3): 393–5. doi:10.1016/0167-4781(91)90183-m. PMID 1830491.
- ^ a b c d "Entrez Gene: ATP5F1A ATP synthase F1 subunit alpha".
- ^ Zong NC, Li H, Li H, Lam MP, Jimenez RC, Kim CS, Deng N, Kim AK, Choi JH, Zelaya I, Liem D, Meyer D, Odeberg J, Fang C, Lu HJ, Xu T, Weiss J, Duan H, Uhlen M, Yates JR, Apweiler R, Ge J, Hermjakob H, Ping P (October 2013). "Integration of cardiac proteome biology and medicine by a specialized knowledgebase". Circulation Research. 113 (9): 1043–53. doi:10.1161/CIRCRESAHA.113.301151. PMC 4076475. PMID 23965338.
- ^ "ATP synthase subunit alpha, mitochondrial". Cardiac Organellar Protein Atlas Knowledgebase (COPaKB). Archived from the original on 2018-07-20. Retrieved 2018-07-18.
- ^ Kagawa Y, Racker E (May 1966). "Partial resolution of the enzymes catalyzing oxidative phosphorylation. 8. Properties of a factor conferring oligomycin sensitivity on mitochondrial adenosine triphosphatase". The Journal of Biological Chemistry. 241 (10): 2461–6. doi:10.1016/S0021-9258(18)96640-8. PMID 4223640.
- ^ Mccarty RE (November 1992). "A PLANT BIOCHEMIST'S VIEW OF H+-ATPases AND ATP SYNTHASES". The Journal of Experimental Biology. 172 (Pt 1): 431–441. doi:10.1242/jeb.172.1.431. PMID 9874753.
- ^ Fernandez Moran H, Oda T, Blair PV, Green DE (July 1964). "A Macromolecular Repeating Unit Of Mitochondrial Structure and Function. Correlated Electron Microscopic and Biochemical Studies of Isolated Mitochondria and Submitochondrial Particles of Beef Heart Muscle". The Journal of Cell Biology. 22 (1): 63–100. doi:10.1083/jcb.22.1.63. PMC 2106494. PMID 14195622.
- ^ "ATP synthase subunit alpha, mitochondrial". UniProt. The UniProt Consortium.
- ^ "ATP5F1A". NCBI Genetics Home Resource.
- ^ Joshi T, Singh AK, Haratipour P, Farzaei MH (2019). "Targeting AMPK signaling pathway by natural products for treatment of diabetes mellitus and its complications". Journal of Cellular Physiology. 234 (10): 17212–17231. doi:10.1002/jcp.28528. PMID 30916407. S2CID 85533334.
Further reading
- Dawson SJ, White LA (May 1992). "Treatment of Haemophilus aphrophilus endocarditis with ciprofloxacin". The Journal of Infection. 24 (3): 317–20. doi:10.1016/S0163-4453(05)80037-4. PMID 1602151.
- Kovalyov LI, Shishkin SS, Efimochkin AS, Kovalyova MA, Ershova ES, Egorov TA, Musalyamov AK (July 1995). "The major protein expression profile and two-dimensional protein database of human heart". Electrophoresis. 16 (7): 1160–9. doi:10.1002/elps.11501601192. PMID 7498159. S2CID 32209361.
- Abrahams JP, Leslie AG, Lutter R, Walker JE (August 1994). "Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria". Nature. 370 (6491): 621–8. Bibcode:1994Natur.370..621A. doi:10.1038/370621a0. PMID 8065448. S2CID 4275221.
- Akiyama S, Endo H, Inohara N, Ohta S, Kagawa Y (September 1994). "Gene structure and cell type-specific expression of the human ATP synthase alpha subunit". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1219 (1): 129–40. doi:10.1016/0167-4781(94)90255-0. PMID 8086450.
- Jabs EW, Thomas PJ, Bernstein M, Coss C, Ferreira GC, Pedersen PL (May 1994). "Chromosomal localization of genes required for the terminal steps of oxidative metabolism: alpha and gamma subunits of ATP synthase and the phosphate carrier". Human Genetics. 93 (5): 600–2. doi:10.1007/bf00202832. PMID 8168843. S2CID 39597611.
- Godbout R, Bisgrove DA, Honoré LH, Day RS (January 1993). "Amplification of the gene encoding the alpha-subunit of the mitochondrial ATP synthase complex in a human retinoblastoma cell line". Gene. 123 (2): 195–201. doi:10.1016/0378-1119(93)90124-L. PMID 8428659.
- Godbout R, Pandita A, Beatty B, Bie W, Squire JA (1997). "Comparative genomic hybridization analysis of Y79 and FISH mapping indicate the amplified human mitochondrial ATP synthase alpha-subunit gene (ATP5A) maps to chromosome 18q12-->q21". Cytogenetics and Cell Genetics. 77 (3–4): 253–6. doi:10.1159/000134588. PMID 9284928.
- Elston T, Wang H, Oster G (January 1998). "Energy transduction in ATP synthase". Nature. 391 (6666): 510–3. Bibcode:1998Natur.391..510E. doi:10.1038/35185. PMID 9461222. S2CID 4406161.
- Wang H, Oster G (November 1998). "Energy transduction in the F1 motor of ATP synthase". Nature. 396 (6708): 279–82. Bibcode:1998Natur.396..279W. doi:10.1038/24409. PMID 9834036. S2CID 4424498.
- Moser TL, Stack MS, Asplin I, Enghild JJ, Højrup P, Everitt L, Hubchak S, Schnaper HW, Pizzo SV (March 1999). "Angiostatin binds ATP synthase on the surface of human endothelial cells". Proceedings of the National Academy of Sciences of the United States of America. 96 (6): 2811–6. Bibcode:1999PNAS...96.2811M. doi:10.1073/pnas.96.6.2811. PMC 15851. PMID 10077593.
- Moser TL, Kenan DJ, Ashley TA, Roy JA, Goodman MD, Misra UK, Cheek DJ, Pizzo SV (June 2001). "Endothelial cell surface F1-F0 ATP synthase is active in ATP synthesis and is inhibited by angiostatin". Proceedings of the National Academy of Sciences of the United States of America. 98 (12): 6656–61. Bibcode:2001PNAS...98.6656M. doi:10.1073/pnas.131067798. PMC 34409. PMID 11381144.
- Wang ZG, White PS, Ackerman SH (August 2001). "Atp11p and Atp12p are assembly factors for the F(1)-ATPase in human mitochondria". The Journal of Biological Chemistry. 276 (33): 30773–8. doi:10.1074/jbc.M104133200. PMID 11410595.
- Chang SY, Park SG, Kim S, Kang CY (March 2002). "Interaction of the C-terminal domain of p43 and the alpha subunit of ATP synthase. Its functional implication in endothelial cell proliferation". The Journal of Biological Chemistry. 277 (10): 8388–94. doi:10.1074/jbc.M108792200. PMID 11741979.
- Sergeant N, Wattez A, Galván-valencia M, Ghestem A, David JP, Lemoine J, Sautiére PE, Dachary J, Mazat JP, Michalski JC, Velours J, Mena-López R, Delacourte A (2003). "Association of ATP synthase alpha-chain with neurofibrillary degeneration in Alzheimer's disease". Neuroscience. 117 (2): 293–303. doi:10.1016/S0306-4522(02)00747-9. PMID 12614671. S2CID 43991411.
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G (February 2004). "A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway". Nature Cell Biology. 6 (2): 97–105. doi:10.1038/ncb1086. PMID 14743216. S2CID 11683986.
- Cross RL (January 2004). "Molecular motors: turning the ATP motor". Nature. 427 (6973): 407–8. Bibcode:2004Natur.427..407C. doi:10.1038/427407b. PMID 14749816. S2CID 52819856.
- Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O'Donnell P, Taylor P, Taylor L, Zougman A, Woodgett JR, Langeberg LK, Scott JD, Pawson T (August 2004). "Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization". Current Biology. 14 (16): 1436–50. doi:10.1016/j.cub.2004.07.051. PMID 15324660.
External links
- Human ATP5A1 genome location and ATP5A1 gene details page in the UCSC Genome Browser.
- Overview of all the structural information available in the PDB for UniProt: P80021 (Pig ATP synthase subunit alpha, mitochondrial) at the PDBe-KB.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
- v
- t
- e
-
 1bmf: BOVINE MITOCHONDRIAL F1-ATPASE
1bmf: BOVINE MITOCHONDRIAL F1-ATPASE -
 1cow: BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B
1cow: BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B -
 1e1q: BOVINE MITOCHONDRIAL F1-ATPASE AT 100K
1e1q: BOVINE MITOCHONDRIAL F1-ATPASE AT 100K -
 1e1r: BOVINE MITOCHONDRIAL F1-ATPASE INHIBITED BY MG2+ADP AND ALUMINIUM FLUORIDE
1e1r: BOVINE MITOCHONDRIAL F1-ATPASE INHIBITED BY MG2+ADP AND ALUMINIUM FLUORIDE -
 1e79: BOVINE F1-ATPASE INHIBITED BY DCCD (DICYCLOHEXYLCARBODIIMIDE)
1e79: BOVINE F1-ATPASE INHIBITED BY DCCD (DICYCLOHEXYLCARBODIIMIDE) -
 1efr: BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE PEPTIDE ANTIBIOTIC EFRAPEPTIN
1efr: BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE PEPTIDE ANTIBIOTIC EFRAPEPTIN -
 1h8e: (ADP.ALF4)2(ADP.SO4) BOVINE F1-ATPASE (ALL THREE CATALYTIC SITES OCCUPIED)
1h8e: (ADP.ALF4)2(ADP.SO4) BOVINE F1-ATPASE (ALL THREE CATALYTIC SITES OCCUPIED) -
 1h8h: BOVINE MITOCHONDRIAL F1-ATPASE CRYSTALLISED IN THE PRESENCE OF 5MM AMPPNP
1h8h: BOVINE MITOCHONDRIAL F1-ATPASE CRYSTALLISED IN THE PRESENCE OF 5MM AMPPNP -
 1mab: RAT LIVER F1-ATPASE
1mab: RAT LIVER F1-ATPASE -
 1nbm: THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN
1nbm: THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN -
 1ohh: BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE INHIBITOR PROTEIN IF1
1ohh: BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE INHIBITOR PROTEIN IF1 -
 1w0j: BERYLLIUM FLUORIDE INHIBITED BOVINE F1-ATPASE
1w0j: BERYLLIUM FLUORIDE INHIBITED BOVINE F1-ATPASE -
 1w0k: BERYLLIUM FLUORIDE INHIBITED BOVINE F1-ATPASE
1w0k: BERYLLIUM FLUORIDE INHIBITED BOVINE F1-ATPASE -
 2ck3: AZIDE INHIBITED BOVINE F1-ATPASE
2ck3: AZIDE INHIBITED BOVINE F1-ATPASE -
 2f43: Rat liver F1-ATPase
2f43: Rat liver F1-ATPase -
 2jdi: GROUND STATE STRUCTURE OF F1-ATPASE FROM BOVINE HEART MITOCHONDRIA (BOVINE F1-ATPASE CRYSTALLISED IN THE ABSENCE OF AZIDE)
2jdi: GROUND STATE STRUCTURE OF F1-ATPASE FROM BOVINE HEART MITOCHONDRIA (BOVINE F1-ATPASE CRYSTALLISED IN THE ABSENCE OF AZIDE)